Why choose live-cell imaging
Maybe you ask yourself why you should choose the method of live-cell imaging in the first place. To explore the dynamics of living cells, live-cell imaging is one of the major techniques. It supports your study of the cell behavior towards external stimuli. What sounds so simple in theory is anything but easy in practice. The main challenge for live-cell imaging is to get high-quality images in a minimal time frame with reduced phototoxicity for living cells.
Genetically modified fluorescence tagged proteins
With the discovery of fluorescence proteins like the GFP- green fluorescence protein from the jellyfish Aequorea Victoria, the era of genetically modified fluorescence tagged proteins began. A variety of other fluorescence proteins were designed with different fluorescence emission spectra like RFP (red fluorescent protein) and YFP (yellow fluorescent protein). Their discovery was revolutionary. They literally bring light into the darkness. The genetically engineered fusion of these proteins to other proteins (named GFP-tag) by cloning methods offers the possibility to visualize the temporal and spatial localization of the tagged protein in the cell. But that’s not the only advantage. Let’s combine different fluorescent proteins with multiple spectral profiles. Now it’s possible to study the dynamics of the interaction of different proteins.
Therefore, a need for high-resolution imaging systems of collecting images rapidly, with illumination levels low enough to avoid damaging light-sensitive fluorophores and cells, arises.
3D Imaging
Another difficulty when performing live-cell imaging is the 3-dimensional imaging. It requires optical sectioning of the sample. One possibility to image samples in 3D is widefield fluorescence microscopy. But these images often contain a high level of background signal that obscures specimen detail and dramatically reduces contrast. This problem can be bypassed by deconvolution, a mathematical software that is used to restore an image in order to make it sharper. It still keeps a low frame rate of the system.
Confocal laser scanning microscopy
Confocal laser scanning microscopy offers the possibility to perform live-cell imaging in a high spatial and temporal manner. Single-point scanners that are often used for confocal microscopy sequentially build up an image with the cost of very slow image acquisition.
But through scanning an array of points across the sample and collecting fluorescence emissions through a set of confocal pinholes by a spinning disk (Nipkow disk), increases the acquisition speed of an image through a high frame rate of the system.
Confocal spinning unit (CSU)
Another important aspect of spinning disks is the pinhole distance and size. Reducing the pinhole size and distance may lead to less photobleaching but also less confocality. Whereas increasing may lead to phototoxicity of the laser light. Therefore, the pinhole size and spacing issue is always a tradeoff between generating enough light to successfully image the specimen and maintaining a high degree of confocality. The first versions of this design suffered from the poor transmission of illumination through the disk, especially for dim and thick specimens.
But this problem has been solved by the unique design of Yokogawa’s microlenses enhanced spinning disk that is able to focus 40% of the illuminating light through the pinholes, named the CSU (confocal scanning unit).
Application areas of CSU
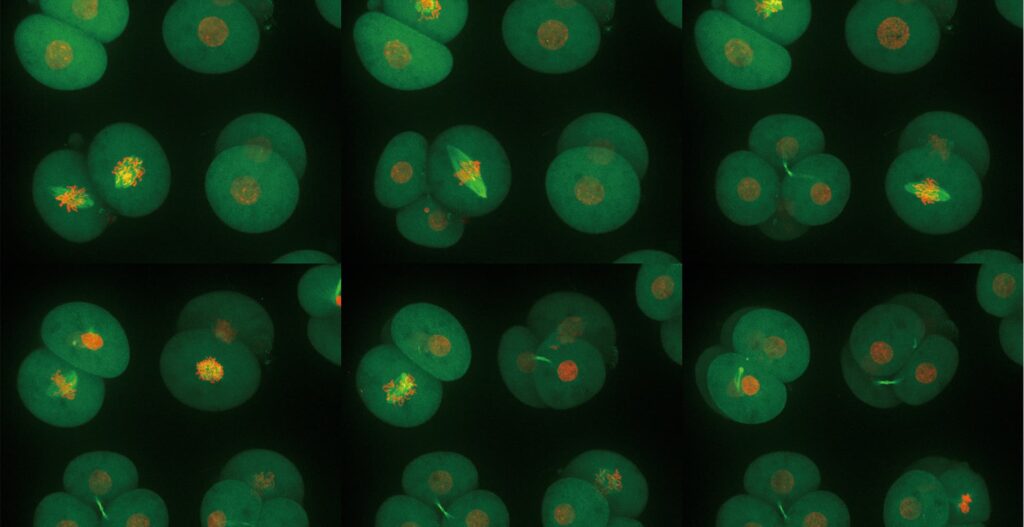
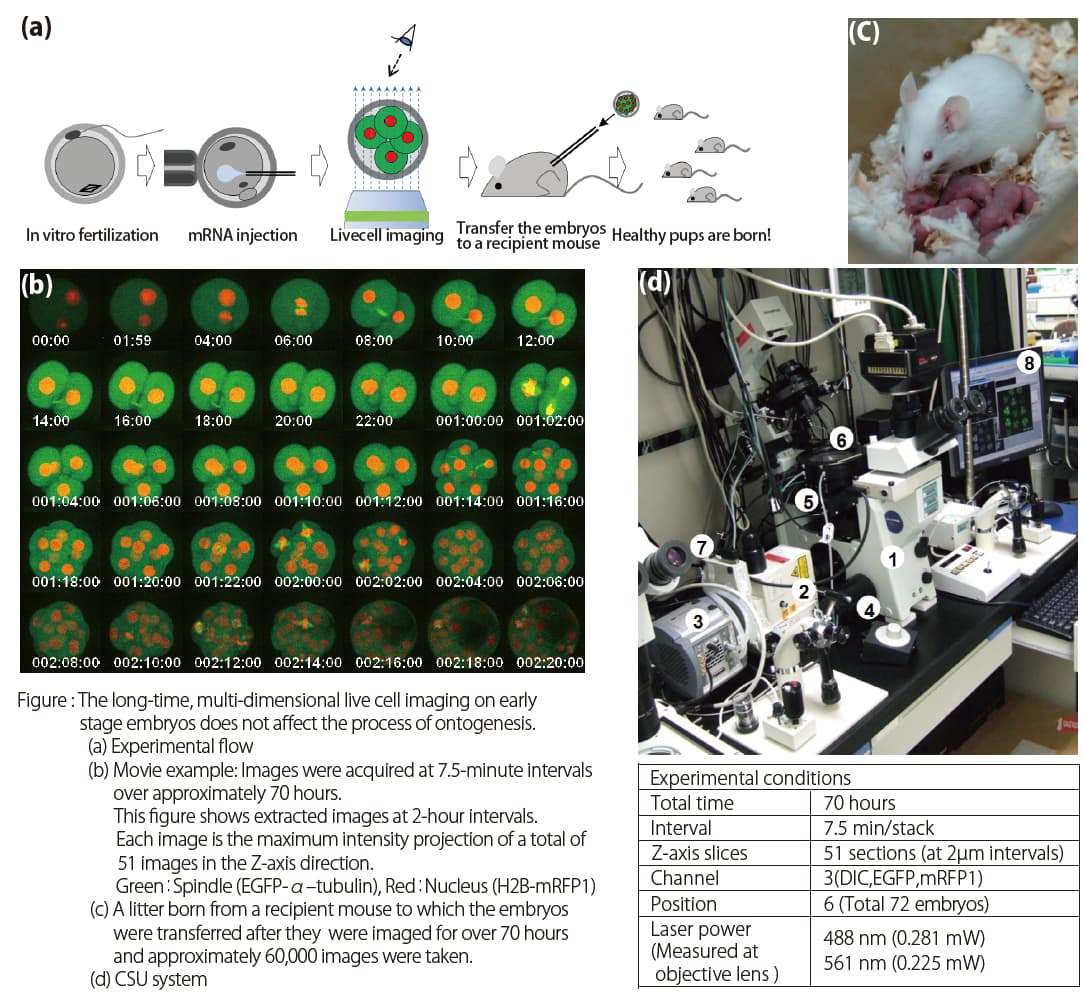
The extremely low phototoxicity of the CSU was proven by long-time, multi-dimensional imaging of early embryonic development of mice by Dr. Kazuo Yamagata. He established such a technique by using the CSU system. He could successfully image mouse embryos over a long period of time. After transferring them to a recipient mouse, the pups were born all normally, grew healthy, and were capable of reproduction (see Figures below).
Another CSU application example leads to the unraveling of the molecular mechanism of pathological conditions of metabolic syndrome by in vivo molecular imaging by Satoshi Nishimura M.D., Ph.D. With this method, it’s possible to precisely evaluate three-dimensional changes in the
structures in living tissue, and the multi-cellular dynamics in vivo with high temporal and spatial resolutions. Even real-time observation of neuronal activity in a zebrafish larva via GCaMP calcium indicator is enabled. With CSU, the brain activity in zebrafish larvae at a remarkably high resolution can be imaged. This was published by Professor Koichi Kawakami and Associate Professor Akira Muto, Department of Developmental Genetics, National Institute of Genetics, Japan.
Even the interface of plants and atmospheric environment published by Takumi Higaki, PhD. and Professor Seiichiro Hasezawa, Dept. of Integrated Bioscience, School of Frontier Sciences can be captured by live imaging.
Why CSU makes your lab life easier
In summary, our CSU offers the possibility to perform live-cell imaging with less phototoxicity at high speed without loss of confocality through the microlenses enhanced spinning disk. Wouldn’t it be great to get better pictures in less time?
Read here our interview with imaging expert Asako Sugimoto, Ph. D.
What are common obstacles you experience while live-cell imaging? Share your experiences with us in the comment section.