For more than one year the world is struggling with severe acute respiratory syndrome (SARS) caused by a single-stranded RNA pathogen SARS coronavirus 2 (SARS-CoV-2). In March 2020, the World Health Organization (WHO) categorized COVID-19 disease as a pandemic. SARS-CoV-2 can cause acute and chronic respiratory and central nervous system illnesses (1,2). How can researchers use genomic sequencing to get more insights into the deadly virus?
Entry of SARS-CoV-2
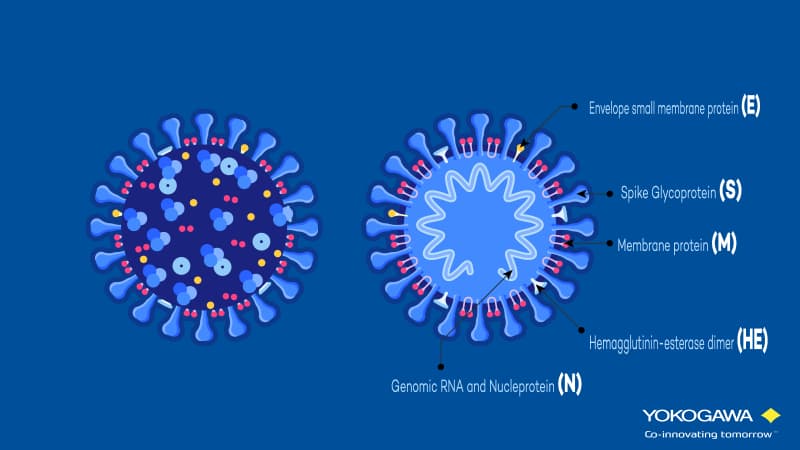
The entry of SARS-CoV-2 occurs through the Angiotensin Converting Enzyme (ACE) 2 receptor. It is expressed on the cell surface of different cell types, especially in the lung. What does the virus there? It is binding with its receptor-binding domain (RBD) of the spike protein (see figure below). This leads to viral entry and replication, as well as severe lung injury.
Developing a vaccine
Based on this finding vaccines were developed against SARS-CoV-2 which consist of the coding sequence for the spike protein either in form of a vector (DNA), developed by AstraZeneca and Johnson & Johnson, or in form of an mRNA (messenger RNA) developed by Biontech and Moderna. But one problem which recently came up is the high mutation rate of SARS-CoV-2. The mutations are mainly occurring in regions of the spike protein which can possibly lead to the emergence of escape strains that are partially or fully resistant to existing vaccinations. RNA viruses have a high mutation rate due to frequent recombination that can produce new genotypes.
Using genomic sequencing for tracking of different mutant strains
The novel SARS-CoV-2 variant was first discovered in the United Kingdom on 20 September 2020. This variant is a rapidly growing lineage with a high replicative advantage, leading to faster spreading of the virus (3). Therefore, it is crucial to track mutations that arise in this strain via genomic sequencing, which could further increase its replicative advantage. Tracking of different mutant strains began throughout the whole world via genomic sequencing. Finding genomic alterations in different virus strains is crucial for the selection of new epitopes for drugs or vaccines. Since the whole-genome was sequenced for the first time, about 50.000 SARS-CoV-2 genomic sequences have been submitted to NCBI Nucleotide records and Nextstrain database (4,5).
[vc_cta h2=”Single Cellome™ Unit SU10 Research Grant Program 2021″ add_button=”bottom” btn_title=”Apply today!” btn_color=”orange” btn_link=”url:https%3A%2F%2Fcp.yokogawa.com%2FSU10-Research-Equipment-Grant.html%3Fid%3Dblog|target:_blank”]Would you like to work with the Single Cellome™ Unit SU10 for 6 months for free? Would you like to present your result results at a Yokogawa webinar? Apply for our SU10 Research Equipment Grant Program 2021!

The importance of genomic sequencing for drug discovery
I think it’s crucial and a blessing to be able to use genomic sequencing. It makes the decisive step from guessing into analyzing the virus. The better you know the virus and its genomic sequence, the better you’ll be able to find matching drugs and active agents. With genomic sequencing, the whole research process is made more efficient and faster.
[vc_separator color=”blue”]
Resources:
- To KK, Hung IF, Chan JF, Yuen KY. From SARS coronavirus to novel animal and human coronaviruses. J Thorac Dis. 2013;5(Suppl 2):S103–S108. [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung SH. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59:6595–6628. [PMC free article] [PubMed] [Google Scholar]
- Grabowski F, Preibisch G, Giziński S, Kochańczyk M, Lipniacki T. SARS-CoV-2 Variant of Concern 202012/01 Has about Twofold Replicative Advantage and Acquires Concerning Mutations. Viruses. 2021; 13(3):392. https://doi.org/10.3390/v13030392
- Hadfield J, Megill C, Bell SM, et al. Next strain: real-time tracking of pathogen evolution. Bioinformatics 2018;34:4121–3.
- Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265–9.