The fields of single-cell proteomics and metabolomics are advancing very rapidly.
The workflow for proteomics includes:
- sample isolation and preparation
- liquid-phase separation and ionization
- gas-phase separation
- mass spectrometry
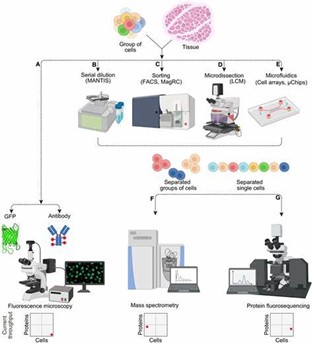
Major techniques used for the isolation of individual target cells for single-cell proteomics are FACS (fluorescence-activated cell sorting) or flow cytometry, laser microdissection, manual cell-picking (micromanipulator), random seeding/dilution, microfluidics/lab-on-a-chip device (Figure 1B-E).
Proteomics: Work Steps
After isolation of the sample preparation, workflows for bottom-up proteomics aim to quantitatively extract proteins from isolated cells or tissues. Proteomics chemically and enzymatically process the extracted proteins to generate peptides and deliver the peptides to a separation platform in ready-to-analyze form and in sufficient quantities to enable a robust measurement. The problem with sample preparation is the high amount of sample loss which even increases for small samples up to 89% and even more for single cells. Therefore, reducing sample processing volumes to avoid as much surface contact as possible is crucial.
nanoPOTS
One platform that deals with that problem is the nanoPOTS (Nanodroplet Processing in One pot for Trace Samples) platform. This platform enables the control of the sample handling conditions based on a robotic handling system for nanoliter volumes. The new technology allows achieving analytical results from small samples that provide orders of magnitude more information than existing technologies. Most samples comprise between 10-14 cells, therefore lacking single-cell resolution.
The following downstream analysis after isolation and sample preparation can be performed by Mass spectrometry or Protein fluorosequencing (Figure 1 F and G).
Spatial metabolomics studies of single cells
Despite single-cell proteomics, Mass spectrometry analysis can be also used in the field of spatial metabolomics studies of single cells. Single-cell metabolomics studies face different challenges such as low picoliter volume of material available for analysis, and large diversity in metabolite structures, properties, and concentrations (from nanomolar to millimolar). Other limitations include co-detection of isomeric and isobaric species, quantification, throughput, and high limits of detection due to low ionization efficiencies of some metabolites and signal division into several mass channels from natural adduct formation.
Metabolomics: Sampling and Ionization
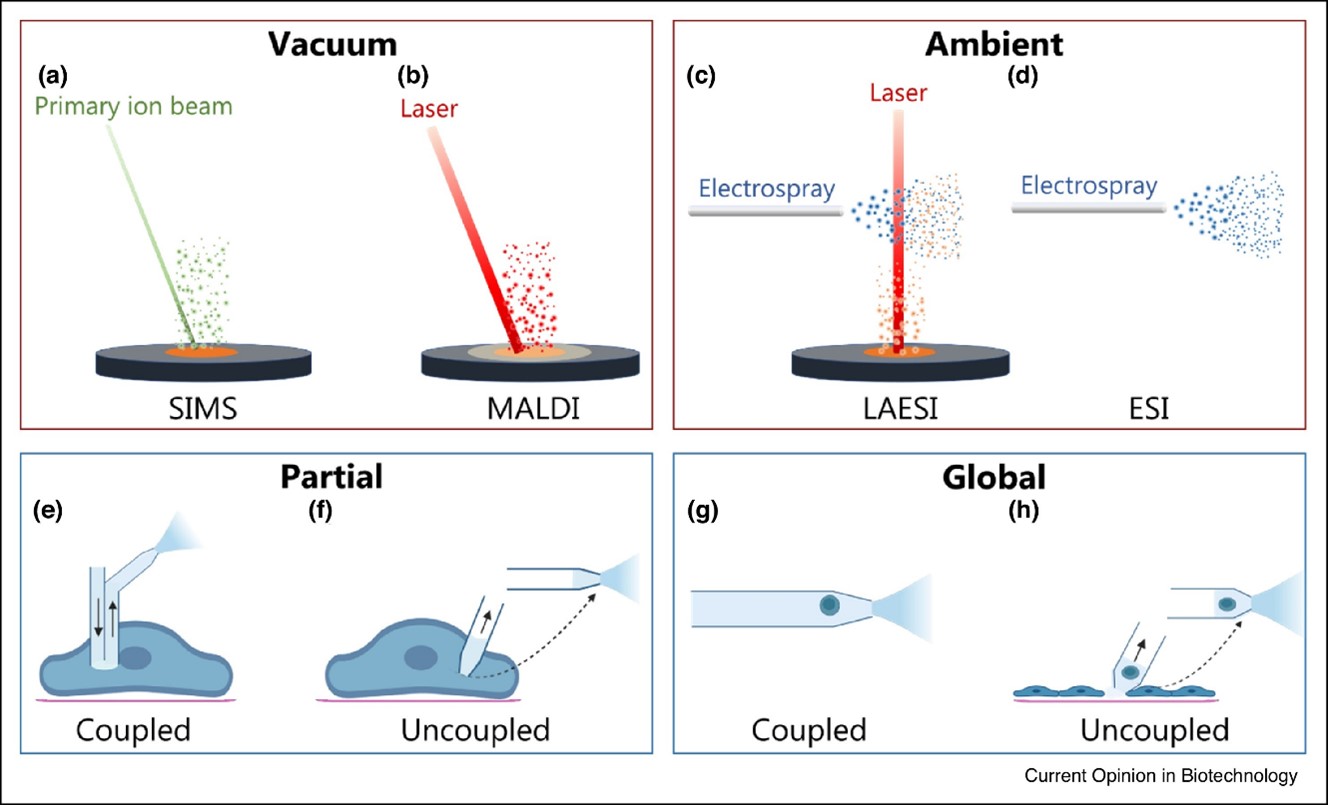
The strategies used for sampling and ionization can be divided into techniques where metabolites are sampled and ionized in the gas phase directly from the solid cell surface such as (Figure 2):
- solid surface with secondary ion mass spectrometry (SIMS)
- matrix-assisted laser desorption (MALDI)
- laser ablation electrospray ionization (LAESI)
And techniques that require metabolites to be sampled into a liquid:
- electrospray ionization (ESI) either by directly infusing the sample or after separating it with capillary electrophoresis (CE) or liquid chromatography (LC)
The most used technique is ESI where molecules are ionized in the liquid phase under high voltage and transferred to gas-phase ions from fine droplets. The choice of voltage, solvent, and flow rate influences the ionization of molecular species.
Strategies for sampling metabolites from individual cells for ESI can be divided into (Figure 2e-h):
- partial sampling where a portion of the cytoplasm is physically removed with a probe or thin capillary
- global sampling where metabolites from the entire cell are desorbed into solution
Transferring the Metabolites
The metabolites are transferred for ESI either directly using a coupled approach or in a two-step fashion using an uncoupled approach. A partial sampling strategy can provide specific chemical information from organelles and the cytosol to target metabolites inside the cell and molecules transported through the plasma membrane. What are the limitations of partial sampling? It takes some time to precisely insert the tip into the cell. Further, the sampling reproducibility is restricted due to the chemically heterogeneous cellular interior.
The partial coupled Strategy
The partial coupled strategy uses a probe with a continuously flowing solvent which is inserted into an adherent cell and the material desorbed into the solvent is directly transported for ESI (Figure 2 e). In contrast to that, in the partial uncoupled strategy, a thin capillary is inserted into an adherent cell to withdraw a small volume of the cytoplasm. Subsequently, the capillary is moved to the MS inlet for ESI (Figure 2f).
Global Sampling
Global sampling for ESI of suspended cells (Figure 2g and h) enables the entire metabolome of the cell to be analyzed in a high throughput analysis, but the limitations include morphological and phenotypical alterations of adherent cells forced into suspension.
Global coupled Strategy
The global coupled strategy (Figure 2g) is achieved by directly infusing a diluted suspension of cells or separating a cell in a droplet in front of or inside a capillary for ESI. A solvent with high organic content is generally used to simultaneously lyse the cell and desorb the cellular metabolites for subsequent ESI.
Uncoupled sampling strategy
For the uncoupled sampling strategy (Figure 2h), the cell can be adherent on a substrate or trapped in a microwell. The metabolites are then desorbed into a dispensed solvent droplet that is withdrawn into a capillary and moved for ESI, or the entire cell is sucked into the capillary for ESI.
Strategies for the Future
Future strategies shall ensure cell integrity to avoid analyzing the metabolome of stressed or dying cells, which is a risk when living cells are dried or fixed and adherent cells are in suspension. And they shall facilitate high throughput sampling and analysis to enable statistical separation of individual cells and subpopulations of cells. With our newly developed system SS2000 for automated single-cell sampling and intracellular component sampling, this can be achieved due to its automated fashion. The micropipette tip can be either used to sample intracellular components or single cells. The tip can be transferred directly into the MS with minimizing the probability of sample loss.

Would you like to try the SS2000 for several months for free? Are you a scientist based in
Europe? Then apply for our SS2000 grant program. The deadline is September 30, 2022.