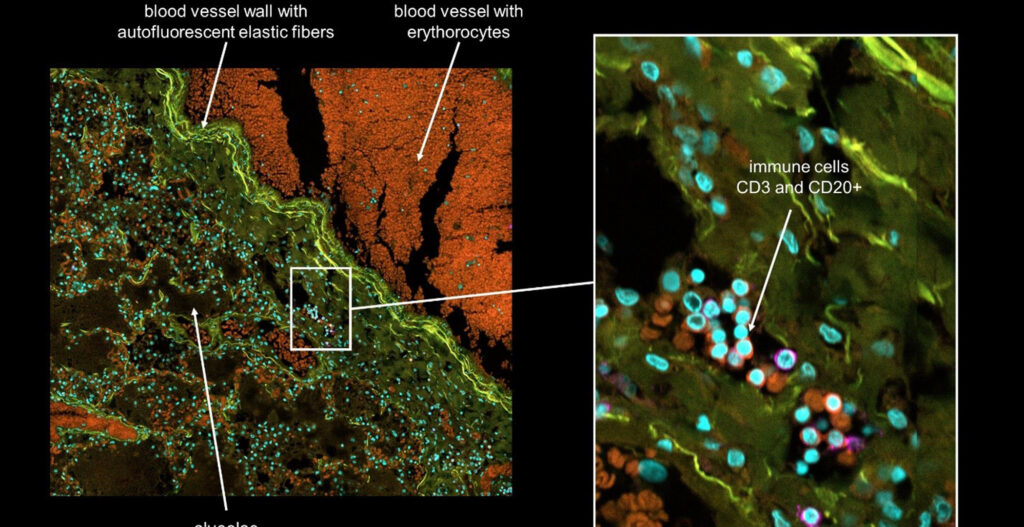
New technology for isolation of single cells and intracellular components
The need for new technologies that enable the analysis of single cells is increasing since last years and includes various fields of life sciences and biomedical research. In most cases, the analysis of heterogeneous cell populations loses important information about a small but potentially relevant subpopulation. This is why our R&D team is currently working on developing a subcellular sampling system.
Single-Cell Isolation Technologies
A variety of single-cell isolation techniques already exist such as FACS (fluorescence associated cell sorting), laser capture microdissection, and microfluidics.
But the most important information is the spatial context of the cell. Unfortunately, it gets lost through FACS and microfluidics technologies because the tissue needs to be dissociated.
Whereas for laser capture microdissection the spatial context is not lost but the cells are damaged through the utilized laser.
This raises the question: How can we isolate single cells while keeping the spatial context intact?
The need for a Subcellular Sampling System
We have news from our innovation team. Currently, our R&D colleagues are developing a subcellular sampling system that enables live cell confocal microscopy imaging and single-cell or intracellular components sampling.
The upcoming subcellular sampling prototype system combines a confocal microscope with powerful analysis software capabilities. Based on acquired images a target list can be defined manually or automatically. An XYZ stage robot picks the identified and selected targets, which could be whole cells or parts of cells. The collected samples can be stored at growth conditions for further use or at low temperatures to avoid degradation.
Confocal microscope of the Subcellar Sampling System
With the confocal microscope, slice images can be acquired at high resolution and 2D and 3D phenotypic characteristics can be obtained before cell sampling. With the incorporated XYZ stage robot, cells can be visualized in real-time during automated single-cell sampling.
These cells can then be analyzed for example by mass spectrometry to measure endogenous or exogenous small molecules of interest. Liliana Pedro and Patrick J. Rudewicz (2020, Anal. Chem.) could demonstrate the potential of using multidimensional and multimodal information collected from single cells to build predictive models for cell classification.
https://youtu.be/uhx46u-BbU8″ title=”Application example: Intracellular content extraction from HepG2 cells
Spatial Omics
Spatially resolved omics technologies have emerged during the last years and have undoubtedly changed the way we understand the spatial organization of complex multicellular biological systems. Spatial and Single Cell Biology platforms cover cutting-edge technologies for spatial profiling of transcripts, proteins, DNA, and small molecules, as well as several approaches for single-cell analysis.
Voice of Customer

We are always looking for innovative and passionate researchers who’d like to test and work with our prototypes. In this case, Professor Melanie Bailey from the University of Surrey tested this prototype over several months. At the NextGen Omics, 04-05 November 2021 in London, she will hold a live talk sharing her experiences and her research focus. Will we meet you in London? For those who can’t be there – don’t worry, the talk will be streamed live.
Explore the possibilities of subcellar sampling!