This blog post highlights the COVID-19 and influenza virus research of MD Ph.D. Yohei Yamauchi and his working group, University of Bristol, United Kingdom. Their main focus is exploring virus entry.
Focusing on Virus Entry
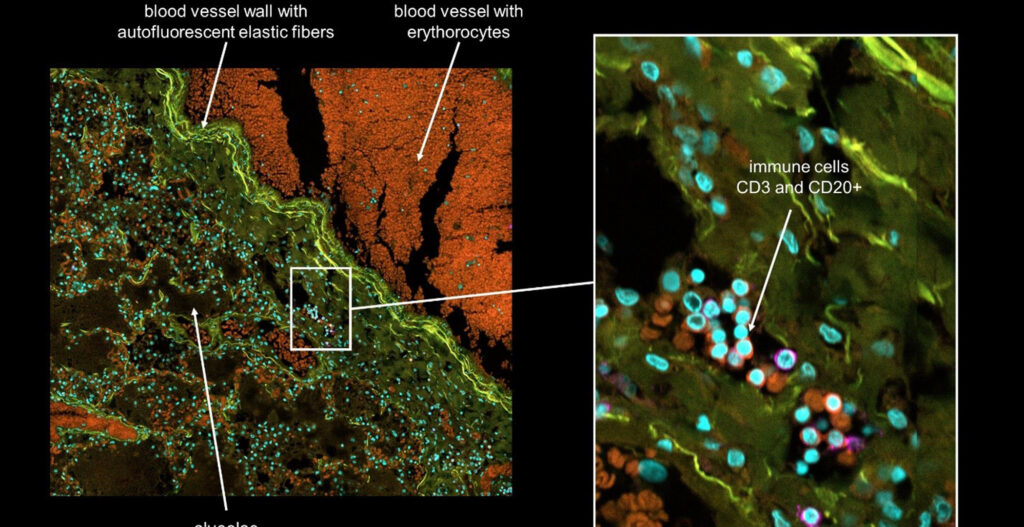
Our group uses automated microscopy and quantitative image analysis to understand how human viruses that cause disease such as influenza A virus (IAV) and SARS-CoV-2 (the causative agent of COVID-19) enter, infect and replicate inside the cells in our body. There are over 700 trillion cells in the human body, some of which can be infected with a pathogenic virus and cause disease. In particular, we are curious about “virus entry” – a stepwise process that involves discrete interactions between the incoming virus particle with a cell. The size of a single virus compared to a single cell, is similar to that of a football compared to a football pitch.
Challenges in virus research
There are more viruses than stars in the universe, and over 200 virus species are known to infect humans. Viruses come in various shapes and sizes, but they have one thing in common – they depend on cellular functions to survive and reproduce. What if there are universal mechanisms of virus interactions with the host? Then we can take advantage of such “interactions” and target them by antiviral strategies. However, the challenge is to understand virus-host cell interactions at a mechanistic level because they can be transient and hard to detect. Microscopy is a great tool to tackle these questions.
Using CellVoyager CQ1 to explore virus entry

In our group, we use the Yokogawa CQ1 to capture viral and cellular protein expression in cell culture using imaging plates. These are mostly done using 96-well optical plates. The cells are typically treated with drugs, siRNAs, CRISPR/Cas9, etc. to block a cellular gene or function. The cells are infected with a virus and the infection phenotype (reduction or increase) is imaged at different stages of infection, followed by automated image analysis supported by machine learning. In our lab, we have established image-based assays to quantitate each and every step of virus entry.
Examples of these steps are:
- attachment
- internalization
- acidification in endosomes
- viral fusion with cellular membranes
- uncoating
- penetration.
Recently we identified the neuropilin-1 receptor of SARS-CoV-2 and showed its importance for viral uptake into cells (Daly et al., 2020; Cantuti-Castelvetri et al., 2020), and identified the impact of non-canonical autophagy in IAV entry (Wang et al., 2021). CQ1 was a powerful tool in enabling these studies in a timely fashion.
Benefits of working with CellVoyager CQ1
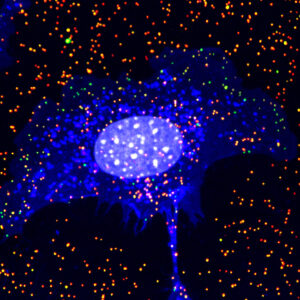
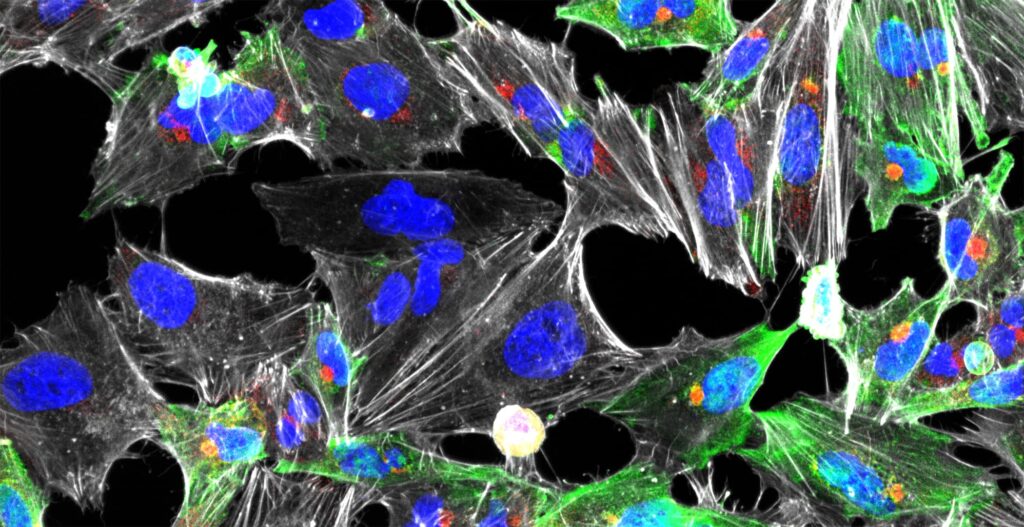
Green, intracellular IAV; Red, extracellular IAV; Blue, What germ agglutinin; Grey, nucleus
CQ1 is compact, reliable, very fast, and easy to use and everyone in my lab loves it. The images are automatically acquired using the spinning-disk confocal, the software is intuitive. The greatest advantage of CQ1 is its incredible speed of image acquisition. Our lab also uses the CellPathfinder created by Yokogawa in order to combine machine learning with image analysis. The Yokogawa team is extremely supportive and helps us in image analysis pipeline generation and trouble-shooting.
Prospects for the future
My lab brings the two realms of cell biology and viruses together to learn from the interface of virus and cell, which is a very exciting and new expanding field. COVID-19 proved that deeper research on virus-host interactions is necessary. Viruses understand their host cells very well. In fact, technologies like the reverse transcriptase used for COVID-19 PCR tests, CRISPR/Cas9 genome editing, gene delivery, etc. all exist as a result of millions of years of interactions between virus and host. Viruses are like locals that have lived in the same city for decades and know every new bar and restaurant and short-cuts to work. On the other hand, researchers are like newcomers who know the main tourist attractions and the road to the airport. Hence viruses are by far the best cell biologists and we scientists can learn a wealth of information from studying their behaviour.
By closely shadowing a virus using microscopy it can lead to unexpected and important cell biological insights. These insights are supported by interdisciplinary research that promotes new discoveries. Some of these findings may potentially lead to novel universal antiviral strategies, that can curb pandemics and tackle emerging viruses of the future. Our group will continue the pursuit of virology combined with cell biology, proteomics, biochemistry, and structural biology and aim for new breakthroughs in virus-informed cell biology.
References
Daly JL et al., Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020 Nov 13;370(6518):861-865. doi: 10.1126/science.abd3072. Epub 2020 Oct 20. PMID: 33082294.
Cantuti-Castelvetri L et al., Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020 Nov 13;370(6518):856-860. doi: 10.1126/science.abd2985. Epub 2020 Oct 20. PMID: 33082293; PMCID: PMC7857391.
Wang Y et al., Non-canonical autophagy functions of ATG16L1 in epithelial cells limit lethal infection by influenza A virus. EMBO J. 2021 Mar 15;40(6):e105543. doi: 10.15252/embj.2020105543. Epub 2021 Feb 15. PMID: 33586810; PMCID: PMC7957399.
Lab website
[vc_separator color=”blue”]
[vc_masonry_grid post_type=”post” max_items=”3″ show_filter=”yes” grid_id=”vc_gid:1633953739701-c7fe59b9-5611-4″ taxonomies=”22″ filter_source=”category”]